The appropriate statistical test is logistic regression. Instead of modeling the probability of the response, logistic regression models the log odds or logit, which equals log(p/(1-p)). This solves various distributional problems and ensures that when the predicted values are converted back to probabilities they will all be between 0 and 1.
We will consider two examples, one with raw data and a continuous predictor variable and one with tabulated data and a categorical predictor variable.
The data are available in the file voting.dat.
> vote <- read.table('voting.dat',header=T)
> names(vote)
[1] "conserv" "voterep"
> attach(vote)
>vote # note that 1 = vote republican, 0 otherwise
conserv voterep
1 1.000 0
2 1.000 0
3 1.143 0
4 1.143 0
5 1.285 0
6 1.429 0
7 1.429 0
8 1.571 0
9 1.714 0
10 1.714 0
11 1.857 0
12 2.143 0
13 2.143 0
14 2.286 0
15 2.286 1
16 2.429 0
17 2.429 0
18 2.429 0
19 2.429 0
20 2.429 0
21 2.429 1
22 2.429 0
23 2.571 0
24 2.571 0
25 2.571 1
26 2.714 1
27 2.714 0
28 2.857 0
29 3.000 1
30 3.000 0
31 3.000 0
32 3.000 1
33 3.000 1
34 3.000 0
35 3.000 0
36 3.143 1
37 3.143 1
38 3.143 0
39 3.143 1
40 3.286 0
41 3.286 1
42 3.286 1
43 3.286 1
44 3.286 0
45 3.429 0
46 3.571 1
47 3.571 1
48 3.571 1
49 3.714 0
50 3.714 1
51 3.714 1
52 3.857 1
53 4.000 0
54 4.143 1
55 4.143 1
56 4.429 1
57 4.571 1
58 4.714 1
59 4.714 1
60 4.857 1
61 4.857 1
62 5.000 1
63 5.000 1
#the appropriate procedure is glm(), generalized linear model, with the distribution
#family specified as binomial
#Note: The "z value" in the following output is normally squared
# and reported as the "Wald Chi-sq" testing that coefficient against zero.
> voteglm <- glm(voterep ~ conserv, family=binomial)
> summary(voteglm)
Call:
glm(formula = voterep ~ conserv, family = binomial)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.0641 -0.6590 -0.1711 0.7189 1.9450
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -6.705 1.759 -3.812 0.000138 ***
conserv 2.177 0.572 3.807 0.000141 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 86.939 on 62 degrees of freedom
Residual deviance: 54.279 on 61 degrees of freedom
AIC: 58.279
Number of Fisher Scoring iterations: 5
#note that the logit (i.e., log odds) increase by 2.177 for each
#unit increase in conservatism score. then the following gives
#the multiplicative factor by which the odds increase for each
#unit increase in conservatism score.
> exp(2.177)
[1] 8.819807
#the following gives the 95% confidence interval for the odds
> exp(confint(voteglm, parm='conserv'))
Waiting for profiling to be done...
2.5 % 97.5 %
3.409245 33.372524
#for logistic regression, there is nothing exactly comparable to R-sq
#however, for binary outcome variables the following provides a
#reasonable approximation
> cor(voterep, predict(voteglm))^2
[1] 0.4071420
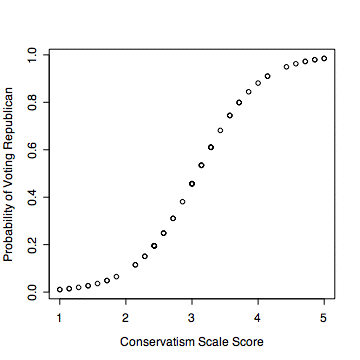
#it is useful to plot the predicted probability from the model
plot(conserv,predict(voteglm,type='response'),
xlab='Conservatism Scale Score',ylab='Probability of Voting Republican')
> water <- factor(c('tap','bottle'))
> well <- c(32, 24)
> sick <- c(49, 6)
> health <- cbind(sick, well)
> vglm <- glm(health ~ water, family=binomial)
> summary(vglm)
Call:
glm(formula = health ~ water, family = binomial)
Deviance Residuals:
[1] 0 0
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.3863 0.4564 -3.037 0.002388 **
watertap 1.8124 0.5099 3.554 0.000379 ***
---
Signif. codes: 0 Ô***Õ 0.001 Ô**Õ 0.01 Ô*Õ 0.05 Ô.Õ 0.1 Ô Õ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 1.5150e+01 on 1 degrees of freedom
Residual deviance: 3.1086e-15 on 0 degrees of freedom
AIC: 12.243
Number of Fisher Scoring iterations: 3
> WaldChiSq = 3.554^2
> WaldChiSq
[1] 12.63092
> exp(1.8124)
[1] 6.12513
> exp(confint(vglm,parm='watertap'))
Waiting for profiling to be done...
2.5 % 97.5 %
2.37829 18.05371
© 2007, Gary McClelland